News Story
The Binding of the Virus
The COVID-19 outbreak has put the world on edge. No country has been untouched as medical teams and policy-makers alike scramble to halt the spread of infection. More than 1.5 million people around the world have died thus far, and the social and economic effects? It’s too soon to tell just how bad they will be. The so-called Spanish Influenza, which extinguished the British tradition of cheek-kissing, has been called both “destroyer and teacher.” From that pandemic, we learned that social distancing and good hygiene greatly reduce the spread of infection, and yet, the numbers of people infected with COVID-19 are beginning to rise again. Inoculations are desperately needed.
A research team in the University of Maryland (UMD) Department of Chemical and Biomolecular Engineering (ChBE) has been looking into why this coronavirus has such a high transmission rate. The team, led by ChBE Professor, Jeffery Klauda, revealed exactly how COVID-19 binds with the receptor protein angiotensin converter enzyme, or ACE2. Additionally, they determined how COVID-19 differs from its cousin, severe acute respiratory syndrome coronavirus (SARS-COV), which caused the 2002 outbreak and why COVID-19 is more deadly. ChBE Ph.D. Student, Mahdi Ghorbani, served as first author on the study.
"In all coronaviruses, a spike protein on the surface of the virion -- the infectious part of the virus that is responsible for transmission -- helps the virus attach to the host cells in the human body by binding to the ACE2," Ghorbani said. "This binding is followed by the fusion of the viral part of coronavirus to the host cell and replication in the human body. So, the first line of attack of coronavirus is receptor binding."
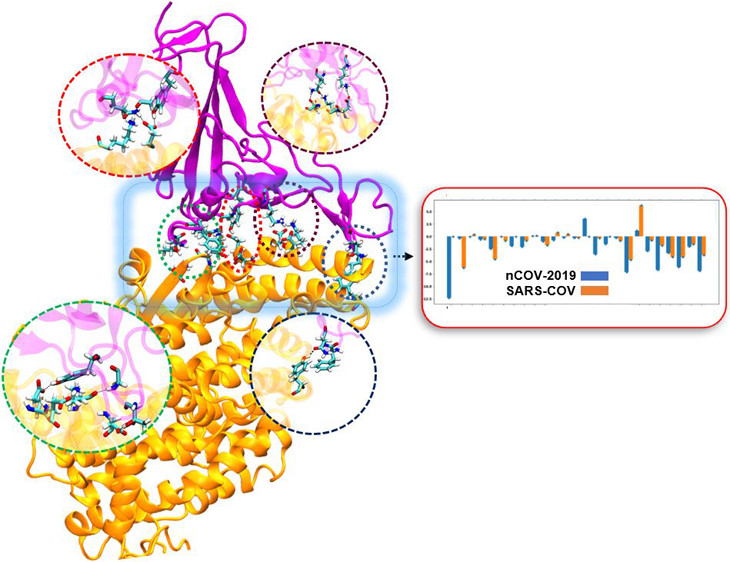
The ability of COVID-19 to attach to ACE2 receptors is an important determinant of viral infectivity in coronaviruses, thus, a major target for vaccination and antiviral strategies. SARS had the same spike protein with roughly 80% similarity to COVID-19, however, COVID-19 is more deadly. This is partly because the receptor binding in COVID-19 is stronger than that of SARS, which leads to a higher rate of transmission.
"Using molecular dynamics simulation and binding free energy calculations, we studied in atomic detail the difference between receptor binding affinity in both COVID-19 and SARS, and found mutations that strengthened the virus-receptor binding" Klauda said.
The team looked into databases for mutations in the receptor-binding domain of coronavirus and found some frequent mutations that were reported all around the world. They studied the effect of these mutations on the binding affinity between coronavirus and the ACE2 protein. Most of these mutations had the same affinity to ACE2 compared with SARS, however, there were a few that increased the binding affinity of COVID-19 to ACE2. These mutations could lead to a stronger and more infectious coronavirus.
"We also performed molecular simulations on the seven most common mutations found from our analysis of receptor-binding domain (RBD) mutations: N439K, T478I, V483A, G476S, S494P, V483F, and A475V," Ghorbani said. "From an evolutionary perspective this study shows the residues in which the virus might further evolve to become even more dangerous to human health.” Moreover, Klauda said, “To our knowledge, this is first detailed molecular simulation study on the effect of various mutations on binding of COVID-19 to ACE2."
The study also addresses the interface of coronavirus and ACE2 via alanine scanning, which conveys the likelihood that a mutation would increase or decrease the binding affinity.
"We found places in the receptor-binding domain of coronavirus where the virus is more vulnerable, and mutation would lead to significantly lower binding affinity," Ghorbani said. "These locations on the coronavirus could be harnessed as a target for drug design purposes and hopefully stop the pandemic."
For additional information:
Ghorbani, Mahdi, Brooks, Bernard R., and Klauda, Jeffery B. "Critical Sequence Hotspots for Binding of Novel Coronavirus to Angiotensin Converter Enzyme as Evaluated by Molecular Simulations." The Journal of Physical Chemistry, vol. 124, no. 45, 2020, pp. 10034-47, DOI:10.1021/acs.jpcb.0c05994
Published December 9, 2020